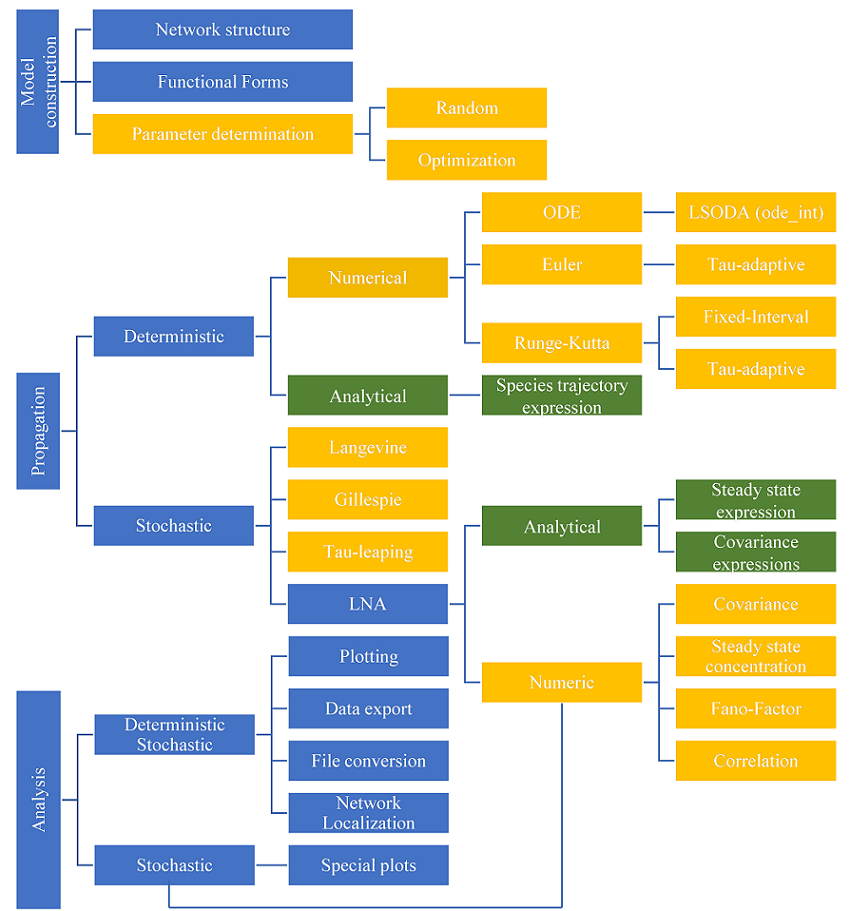
BioSANS is a free software for systems biology which is currently being developed in Academia Sinica Institute of Chemistry. The goal of this development is to make systems biology available to non-domain experts in the easiest possible way. Currently, BioSANS supports model creation, ODE propagation in both deterministic and stochastic settings, and post simulation analysis. It can be used either via the GUI, the command line interface, and as a python import for experts users. BioSANS passed majority of the SBML semnatic and stochastic test cases. It also support parameter estimation and provides an easy to prepare input which follows basic elementary equation in chemistry. In the input file, reaction, initial concentration, and rate constants are required but the propensity expression is optional. The algorithm in BioSANS can infer the propensity from the reactions provided. If the users need special or non mass action type propensity, it can be encoded in the topology file as well. Complicated conditional expression and concentration modification are also supported in the topology file. The following summarized the symbolic and numeric features currently supported;
1. Species analytical expression - works for most linear differential equation and few non-linear ordinary differential equations 2. LNA covariace matrix - works for most linear differential equation and few non-linear ODE 3. Steady state concentration - generally works for most problems especially linear ODE 4. Network localization - topology based sentitivity matrix
1. Linear noise approximation 2. Parameter estimation 3. Network localization 4. Deterministic analysis (ODE integration) * odeint - python LSODA library * runge-kutta4 (tau-adaptive and fixed interval version) * Euler (two differnt types of tau-adaptive version) 5. Stochastic modeling * Chemical Langevine equation (tau-adaptive and fixed interval version) * Tau-leaping algorithm (2 different versions/implementation of Yang Cao's algorithm) * Gillespie direct method
1. Plotting (trajectory, density, etc.) 2. Calculation of correlation, covariance, fano-factor, etc. 3. Phase portrait 4. etc.
Example commands:
propagateA => B, 0.2 &B => C, 0.3whereA=100 &B=0.2 &C=0using CLEwithtn=50 &tlen=1000 &miter=30 &mult_proc=True &fout=Traj1;It can also be type in the console in inline format as followspropagate A => B, 0.2 & B => C, 0.3 where A=100 & B=0.2 & C=0using CLE with tn=50 & tlen=1000 & miter=2 & mult_proc=True & fout=Traj2;
from BioSANS2020 import biosans_lib as biosans# Parameter estimation examplemodelA = """Function_Definitions:Ao = 100Co = 0#REACTIONSA => B, -1B => C, -1@CONCENTRATIONA, AoB, -1C, Co"""dataA = """time,A,C0,100,01,60.65306595,5.7750437622,36.78794412,17.979007163,22.313016,31.827109074,13.5335283,45.001739485,8.208499842,56.5302097310,0.673794694,88.5639249515,0.055308438,97.3057135220,0.004539994,99.3871219425,0.000372665,99.8622879130,3.06E-05,99.9691934435,2.51E-06,99.9931196540,2.06E-07,99.9984642545.5,1.32E-08,99.9997050150,1.38E-09,99.99992352"""if __name__ == '__main__':my_model = biosans.model(modelA).data(dataA)data = my_model.run("k_est6")my_model.clean()